Most of the time, specialist terms used in technical and scientific documents cannot be enumerated in advance. Scientific naming in many domains such as Chemistry, biology, Astronomy, etc. follows nomenclatures and conventions which are highly generative. Similarly, citations to bibliographical items (articles, patents, etc.) present a very high variability. All these expressions are essential to the scientific communication and, as a consequence, to scientific information access tools.
Machine learning techniques are particularly adapted and efficient for recognizing these kind of terms and are used by all our dedicated modules.
Recognition and normalization of quantities
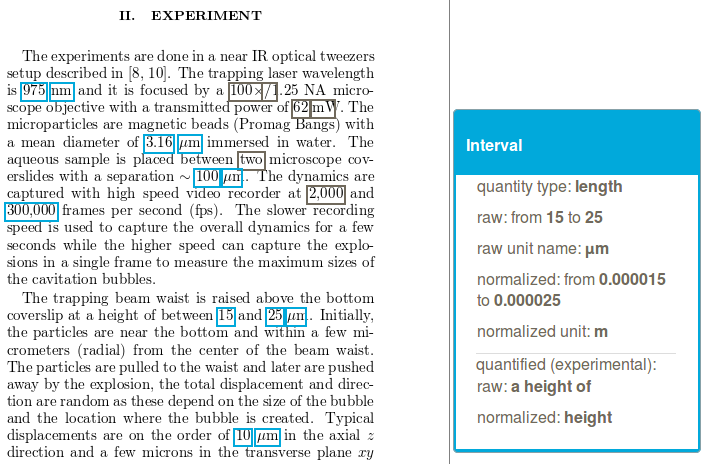
The expressions of quantities and measurement are fundamental in STEM and can be viewed as a particular nomenclature. Identifying and normalizing these expressions into SI base units make possible a vast range of applications, in particular to search quantities in document collections, to apply numerical data mining techniques, or to extract automatically knowledge about experimental conditions.
grobid-quantities recognizes in textual documents (text, PDF, XML) expressions of measurements (e.g. pressure, temperature, etc.), then parses, normalizes them, and finally converts these measurements into SI units. For English, the tool supports more than 120 base units and expressions of atomic, interval and lists of values. In addition, grobid-quantities tries to identify and attached to the measurements the “quantified” substance or objects.
grobid-quantities uses cascades of CRF, similarly as GROBID (therefore the name). It is developed as an Open Source project by science-miner in collaboration with Luca Foppiano (NIMS, Japan).
These normalized measurement can then be exploited by search or data mining tools. Our front-end search tool dedicated to scholar articles for instance allows to express search queries including quantity criteria (e.g. interval) which are then processed with the range query search possibilities of ElasticSearch to retrieve and rank matching documents annotated beforehand by grobid-quantities.
Recognition and extraction of software mentions
Software is today a major form of scientific contribution, however remaining relatively invisible. When software uses are mentioned in articles, due to the lack of standard for software citations, those mentions are often informal. Without possible bibliometrics, the impact of a research software cannot be acknowledged and the incentive of developing and sharing open source research software is today limited for researchers.
In the context of the Softcite project led by James Howison from the University of Texas at Austin and in collaboration with Impactstory, we are developing an Open Source component for recognizing in text and in PDF any mentions of software, together with the associated software attributes such as number version, author, url, version date and related bibliographical citation.
The module performs structure-aware PDF document annotation, exploiting the coordinates of the mentions in the PDF for displaying interactive annotations directly on top the PDF layout, and populating a software knowledge base. Taking advantage of the integration of DeLFT in GROBID, we use a state of the art Deep Learning model (BiLSTM-CRF with ELMo embeddings) and perform the recognition of the software attributes with a f-score currently around 85%.
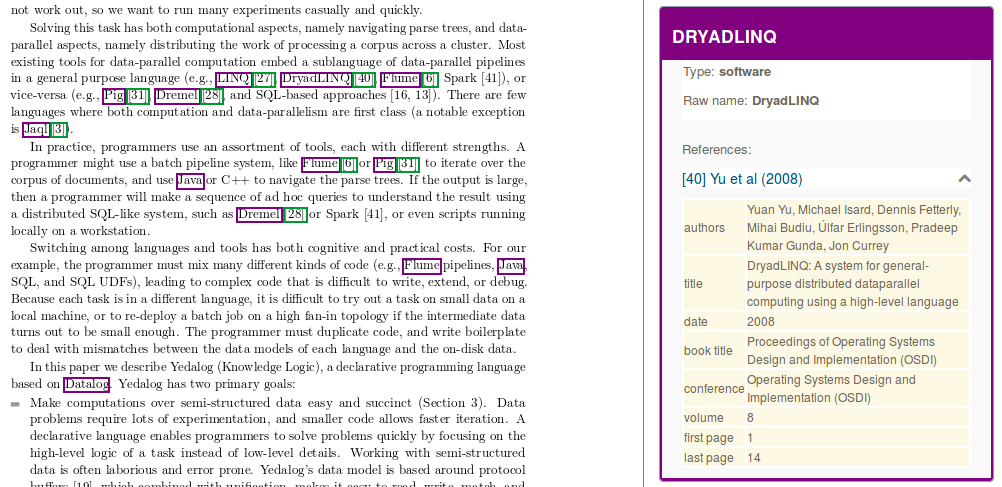
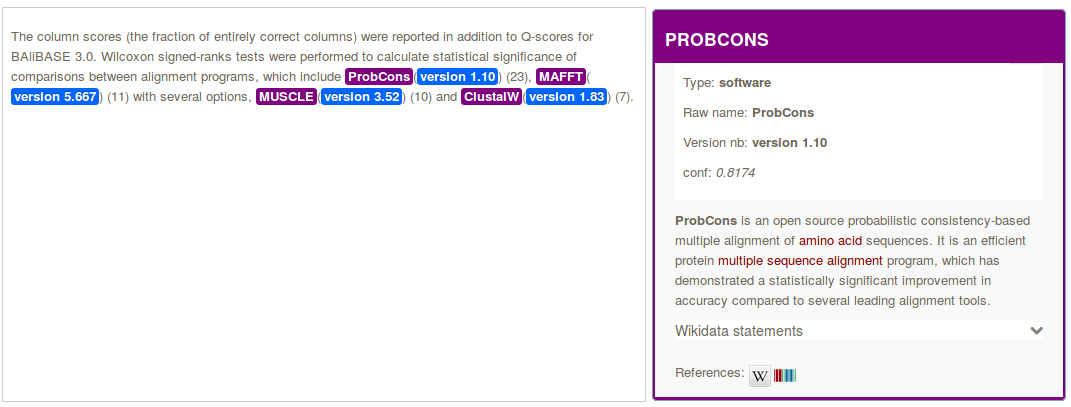
When possible, software entities are disambiguated against Wikidata and associated with the bibliographical references introduced in the context of the mention.